|
|
|
Destockage
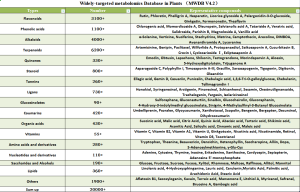
Widely-targeted Metabolomics Database in Rice
|
|
Cette page concerne les importateurs et exportateurs de Widely-targeted Metabolomics Database in Rice Rechercher dans la catégorie : Destockage Rechercher dans la catégorie : targeted, database, metabolomics, widely, rice |
Lundi 28 décembre 2015
Quantité : OEM/ODM Se - Prix : OEM/ODM Service
usinage des pièces de graphite graphite graphite passoire sur mesure produit par notre client, usinage, premier client de fournir leur taille et ensuite choisir le matériel de graphite, nous permettra de fabricant. le graphite passoire a les caractéristiques d'une bonne conductibilité thermique...
XRD Graphite Manufacturing Co., Ltd
- zcxooo
- 467400 - Pingdingshan
- 86 15516002510
Vendredi 31 janvier 2014
Vente des extincteurs et tout matériels pour lutter contre l'incendie; de haute qualité. à un prix défiant toute concurrence. Recharge, Entretien et Vérification de tous les types d'extincteurs. Remplissage et maintenance des extincteurs. **************************************** ******...
Maxi Protection Maroc
- - Adrar
- 00212661343484
Mercredi 14 septembre 2016
Quantité : 5000 - Prix : 50,00 €
Y FILTRE Corps Acier A216 WCB & GS-C25 & 1.0619 & GP240GH - Cartouche inox 304 démontable - Joint Graphite Bouchon de purge acier Montage horizontal ou vertical avec fluide descendant Respecter le sens de passage indiqué sur le corps par une flèche Connection: DIN...
ETM Armaturen
- 59590 - Henri Durre
- 86 577 67953935