Abstract of BGI prenatal test
Objectives
The aim of this study was to assess the performance of noninvasively prenatal testing (NIPT) for fetal copy number variants (CNVs) in clinical samples, using a whole-genome sequencing method.
Method
A total of 919 archived maternal plasma samples with karyotyping/microarray results, including 33 CNVs samples and 886 normal samples from September 1, 2011 to May 31, 2013, were enrolled in this study. The samples were randomly rearranged and blindly sequenced by low-coverage (about 7M reads) whole-genome sequencing of plasma DNA. Fetal CNVs were detected by Fetal Copy-number Analysis through Maternal Plasma Sequencing (FCAPS) to compare to the karyotyping/microarray results. Sensitivity, specificity and were evaluated.
Results
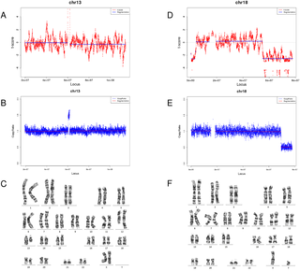
33 samples with deletions/duplications ranging from 1 to 129 Mb were detected with the consistent CNV size and location to karyotyping/microarray results in the study. Ten false positive results and two false negative results were obtained. The sensitivity and specificity of detection deletions/duplications were 84.21% and 98.42%, respectively.
Conclusion
Whole-genome sequencing-based NIPT has high performance in detecting genome-wide CNVs, in particular >10Mb CNVs using the current FCAPS algorithm. It is possible to implement the current method in NIPT to prenatally screening for fetal CNVs.
Introduction
Owing to the discovery of fetal cell-free DNA (cfDNA) in maternal plasma and rapid development of next-generation sequencing (NGS), noninvasive prenatal testing (NIPT) has brought confound changes to antenatal healthcare in the past few years[1]. The clinical validity and utility of NIPT for testing common aneuploidies have been endorsed by various clinical guidelines for using in high risk pregnancies[2]. Future application of NIPT may expand to average–risk pregnancies[2, 3]. However, chromosomal CNVs such as deletion and duplication remain a challenge for NIPT because of their small region of chromosomal abnormality[4]. CNVs are known to commonly exist in human genome, and diseases associated with CNVs, such as DiGeorge syndrome (22q11), Cri-du-chat syndrome (5p-), 1p36 deletion syndrome, are documented[5]. Postnatally, pathogenic CNVs are important contributors of intellectual disabilities in newborns, while in prenatal practice increasing evidence showed that disease-causative CNVs are associated with adverse pregnant outcomes. For instance, in samples with normal karyotype, clinically relevant CNVs were identified in 6% with ultrasound abnormalities and in 1.7% with advanced maternal age or positive serum screening results[6]. Recently, Dong et al showed that among samples referred to chromosomal analysis, 6.4% of samples of products of conception (e.g. spontaneous abortions and stillbirth), 13.5% of prenatal samples, and 26.3% of postnatal samples contained pathogenic CNVs[7]. Unlike aneuploidy, the risk of CNVs in fetus is independent of maternal age, and thus younger pregnant women may equally suffer the risk of pathogenic CNVs as older women[8]. Thus prenatal testing for clinically significant CNVs may bring benefit to clinical management and genetic counseling of pregnant outcome. Currently, amniocentesis or chorionic villus sampling (CVS) followed by karyotyping or microarray is the major approach to identify fetal CNVs, although a small but significant risk of miscarriage is associated with the procedures[9].
Localisation : beijing, 215233 beijing,
Personne à contacter : Ru Tommy , +17 7 22 56 77 42